By Michael Mozdy
Thanks to popular media and movies, when we hear “artificial intelligence,” we might first think of the Terminator or Scarlett Johansson’s voice as the virtual assistant in Her. It's true, some researchers are trying to develop artificial intelligence (AI) that mimics human intelligence (complete with simulated emotion, humor, learning from experience, etc.). But the type of AI most useful to medical science today is the opposite: we use it because it doesn’t operate like human intelligence. It’s faster and it’s not influenced by the same things that influence us.
Perhaps because of the cloud of associations around “artificial intelligence,” Radiology and Imaging Sciences researcher Ed DiBella, PhD, prefers to use the term “deep learning.” The Director of the Utah Center for Advanced Imaging Research (UCAIR), DiBella is cautious about claims that AI is ready to revolutionize medicine right now. His caution stems from a garbage-in-garbage-out concern. All artificial neural networks – what are used in research when we say “deep learning” – need to be trained with a robust set of data (not garbage). “There are several ways this training step can go wrong,” DiBella explains: “if the data set isn’t exhaustive or precise enough, if human error such as a physician’s incomplete diagnosis is included, or if those using the network don’t exactly duplicate the environment of the training data, for instance, the same settings for an MRI scan.”
Yet DiBella believes that there is a subset of AI applications in imaging sciences that can make a big difference right now. “Using deep learning to speed up image reconstruction time is a real game-changer,” he claims, and his group is in the process of publishing three different papers to prove his point.
Clearing Up A Blurry Heart
Tracing the movement of blood within the heart muscle is an important step in determining the extent to which a person might have disease and blockages. But it’s a real pain for imagers – the heart literally jumps around in the chest as it works to pump blood around the body. And that quick movement happens in four chambers in quick succession, so getting a good look is no easy feat.
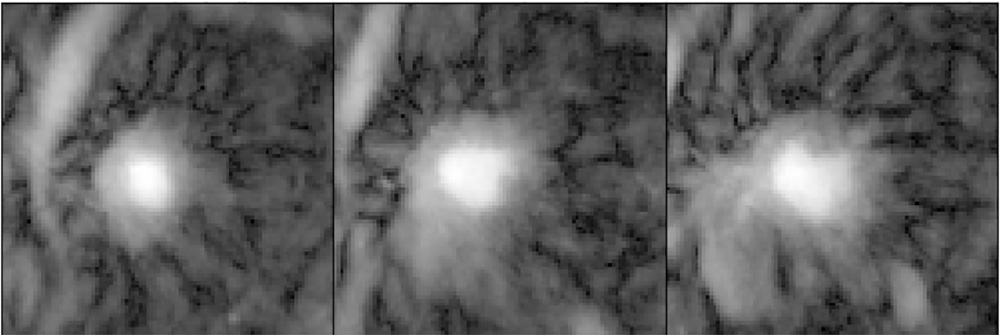
Picture of the sun on a cloudy day? Nope. It’s a cardiac MRI very affected by the motion of the heart.
Cardiologists can choose from several scans, but a SPECT nuclear imaging study is the most used. While SPECT is good at pointing out if there’s a problem area, it takes about 3 hours, it exposes patients to radiation, and it doesn’t have good spatial resolution, meaning cardiologists can’t necessarily pinpoint which exact structures and vessels in the heart are involved (read more about spatial resolution). MRI, on the other hand, provides good spatial resolution as well as a look at the (ir)regularity of the beating and the viability of the muscle tissue. Recently, researchers like DiBella and his colleague Ganesh Adluru, PhD, have overcome problems with MRI perfusion studies related to only acquiring a few “slices” during a normal breath-hold and the resulting artifacts in the images (read about their simultaneous multislice approach). But their solution comes with a significant cost in the time it takes to reconstruct an image from the data they gather: it takes about 2 hours of data crunching, done offsite from the clinical setting, and then the images are sent back to the radiologist or cardiologist.
|
The pretty MRI and CT scans that we see in our doctor’s office are the result of sophisticated algorithms applied to the raw data coming from the scanners. These algorithms apply one or more “filters” to the image. These filters can boost contrast, smooth/sharpen edges, or remove streaks and other artifacts. |
DiBella and Adluru’s solution is something called iterative reconstruction, where a set of partial data (those few slices they acquire during the breath-hold) is slowly tweaked, iteration by iteration, until it results in a sharp image. It requires some baseline knowledge about the way the heart typically looks (some assumptions, that is) in order to successively improve each iteration to a final image. It’s a ton of math and a ton of time, but the results are impressive.
Today, however, off-the-shelf deep learning networks are suddenly available to researchers, and the team saw an opportunity to apply a new tool to this problem. DiBella and Adluru set up their deep learning network to tackle the same iterative reconstruction challenge, only on its own rather than having to run a complex math problem for each data point over and over. Graduate student Johnathan Le was instrumental in their work. Their process:
- Step One: set up the convolutional neural network (CNN) to work with MRI images.
- Step Two: train the CNN with high-quality images (the results from their slow iterative reconstructions). They used 12 data sets from 7 people for this.
- Step Three: test the network by giving it incomplete data and compare the resulting AI images against the ground truth of what those same scans look like from a full data set (which they captured earlier).
Let’s linger on Step Two, Training Time, for a moment to get a sense of how exactly deep learning works. Imagine for a moment that you’re a newly made image. You’re pretty blurry, pretty poor quality because you came from the incomplete data from a fast MRI scan. You were made when Ed Dibella’s team performed a basic mathematical transformation on that data (a Fourier transformation for all you mathletes). The good news is that DiBella’s team ushers you over to a little arena to get you into shape.
The arena (the CNN) is shaped like a U with a set of stairs leading down and a set of stairs leading back up the other side. It turns out that they’re going to make you run down and up these stairs a bunch of times. With each step down, you work out with some weights. In this arena, each weight is just a number, initially set up as a random number around 0 (positive or negative). These weights form filters that will adjust the intensity of your pixels, lighter or darker. This workout on the first step creates 64 copies of you, one for each of the filters that you work out with. It seems pretty random at first, but you’re seeing some potential. Before you move down to the next step, a nifty math equation called downsampling cuts your size in half – every one of the 64 copies of you goes from 144x144 pixels to 72x72 pixels. (“This encodes more information into what is called ‘latent space’,” explains Le, and it’s a common technique in CNNs that helps to reduce training time.)
You see a few more steps where this weight-training will be repeated until you arrive at the bottom of the arena, a slim 18x18 pixel image (512 slightly different copies of you, thanks to all that work with the weights). Now it’s time to run the stairs back up. The weight training happens again, but on this side, an upsampling function combines the copies rather than creating more copies. You see the copies reduce by half with each stair, but you also see yourself filling out, doubling your number of pixels with each stair. When you reach the top, you’re back to just one 144x144 image, and how do you look? Well, not great, but that’s not the point. It turns out DiBella’s team has a good-looking version of you that they kept in reserve (the “ground truth”) and they pull it out for comparison (not to make you feel bad). The arena measures the difference between you and the ground truth, pixel by pixel, then adjusts the weights accordingly, making some heavier and some lighter. Then it’s time for you to run some more stairs, which you’ll continue to do until the result is a good looking image.
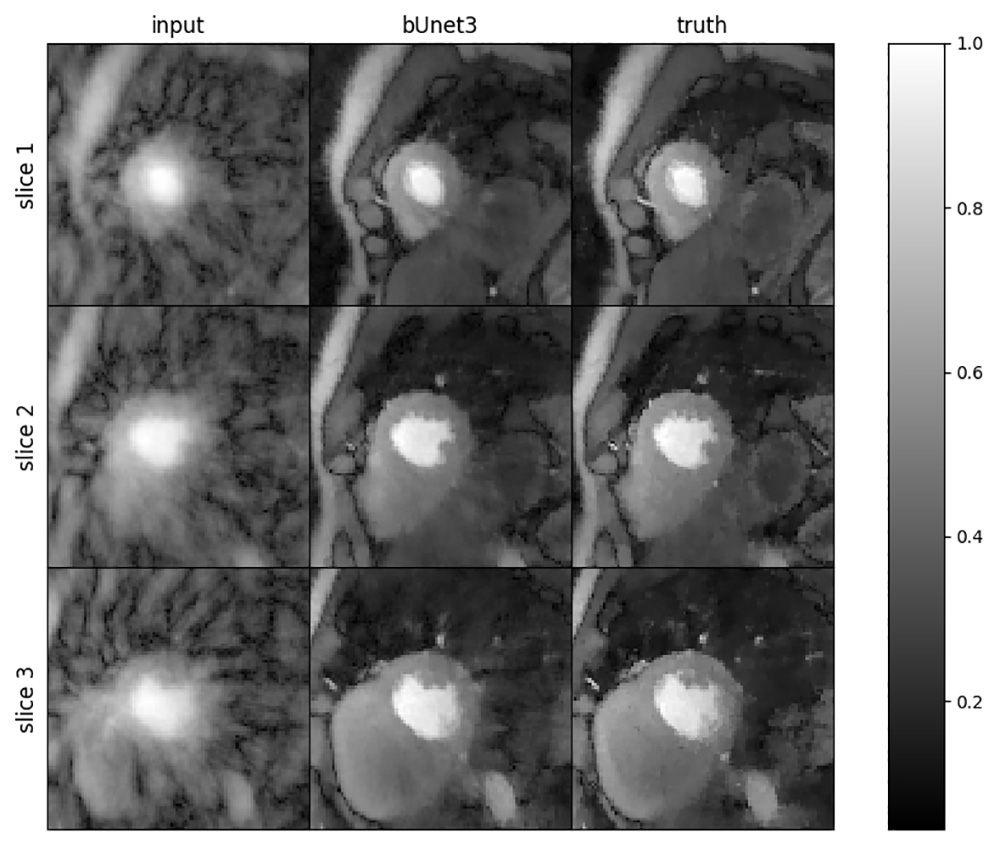
Results of the deep learning algorithm. On the left are slices of the blurry heart without any reconstruction filters. In the middle is the 8-second deep learning approach and on the right is the ground truth as derived from their 30-minute reconstruction algorithm.
Of course, this is a bit simplified. In fact, the network trains on 32 timeframes of the image at once, not just one image. The CNN is made more robust is feeding it many different MRI images of the heart, each one from a different time point or patient, at stress or at rest. The full training set was actually 8,000x2x144x144, or 331,776,000 pixels! Below is a schematic of the CNN architecture (the “arena”).
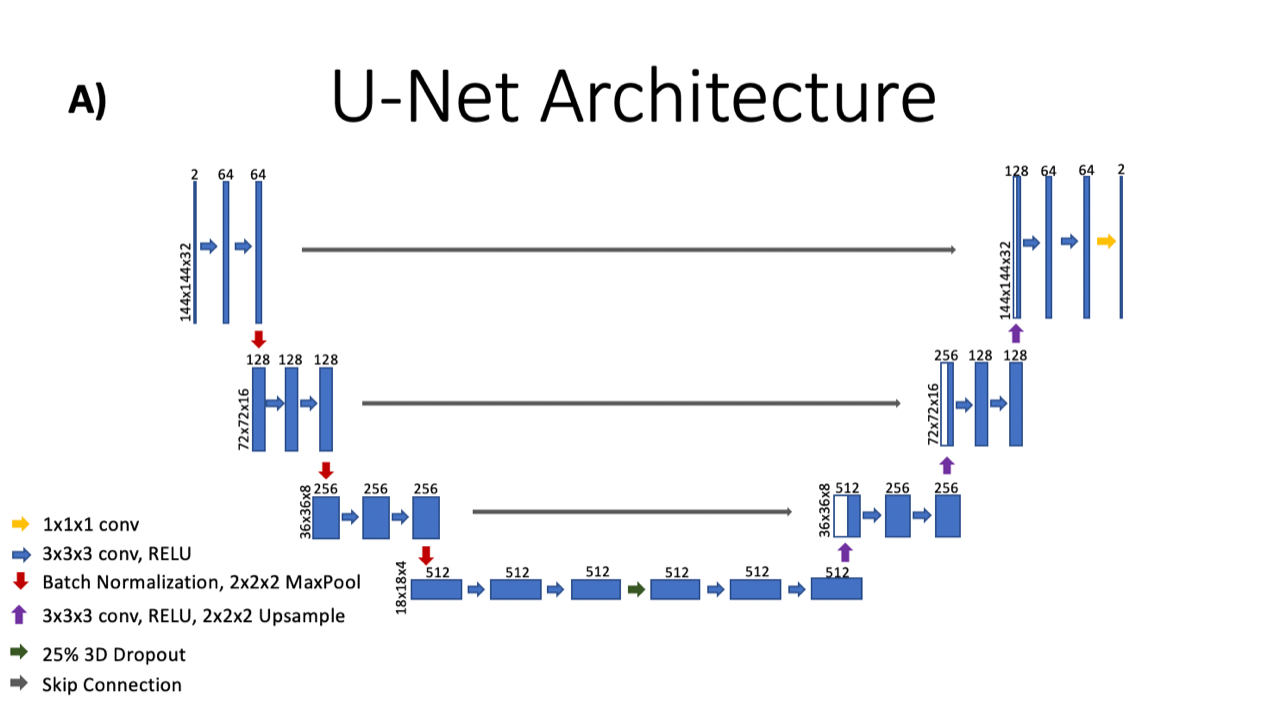
It took Le a full 24 hours to complete the training phase, where the network crunched through the 8,000 images (12 different sets of rest and stress cardiac images). After successful testing, they discovered that this CNN performed similarly to their time-consuming iterative reconstruction. But the time gained with the deep learning technique was significant: to reconstruct just one stress or rest dataset (about 1000 images), the network takes about 8 seconds and the iterative reconstruction takes about 25-35 minutes.
Portability Into the Clinical Environment
University of Utah Health uses MRI scanners manufactured by Siemens, and they’re getting into the AI game, too. Siemens now provides an interface for researchers to upload custom deep learning applications directly on the computer attached to the scanner. Generally, deep learning networks need some serious computing power for the training phase, specifically a GPU that allows for thousands of operations to be completed at once. Once trained, however, they perform well on a more standard CPU. Siemens and other manufacturers take advantage of this fact and make deep learning applications possible right in the clinical environment.
Another paper soon to come out from DiBella’s group describes an application that has already been ported over to the clinical environment. Using a slightly simpler deep learning network called Multi-layer Perceptron (MLP), Adluru is speeding up the scan time for something called T1 mapping of the heart.
|
T1 mapping has many useful applications, notably diagnosing fibrosis in the heart muscle, the liver, and potentially many other organs. It is also a great example of a new direction for MRI: producing quantitative maps of data, not necessarily pictures of structural anatomy. In other words, clinicians are learning to diagnose disease not based on how something looks, but on the tissue values that MRI provides (like T1, T2*, proton density, and magnetization transfer saturation). |
T1 is a measure of how quickly the protons in tissue “relax” back to spinning in a specific direction after an MRI pulse is sent into the body. T1 measurements provide a handy way for clinicians to diagnose coronary artery disease by measuring changes between rest and stress states. The T1 values in regions of the heart (T1 “maps”) at rest provide the baseline. Then a patient is put into a stress state, usually with a drug like adenosine, and T1 maps are made again. Stress makes vessels dilate and more blood is pumped through them, except in the case of blocked arteries, where blood can’t get through. So, healthy vessels show a large change in T1 values (a denser volume of blood cells equals longer T1 times) but diseased arteries show little change in T1 values.
T1 mapping of the heart requires a patient to hold his or her breath for 11 heartbeats (around 11 seconds), but with this new deep learning application, Adluru has cut this down to 5 heartbeats, resulting in an equal quality of image. This is a huge deal for people with coronary artery disease when measuring the stress state: it’s very hard for them to hold their breath for a long time.
A simple analogy for the way Adluru’s model works is to think of different doctors diagnosing a patient. The traditional MRI scan is like a medical resident, freshly graduated from medical school, when a patient shows up with a hurt finger, the resident might need six or seven data points to diagnose it: the patient’s complaint, physical appearance of the finger, checking for signs of broken skin or punctures, ability to flex, tenderness, multiple visual angles of the finger, etc. The T1 mapping scan aided by deep learning is like a veteran orthopedic specialist who has had 50 years of experience (this is the network’s “training”). This expert physician can take in maybe only three data points and make an accurate diagnosis.
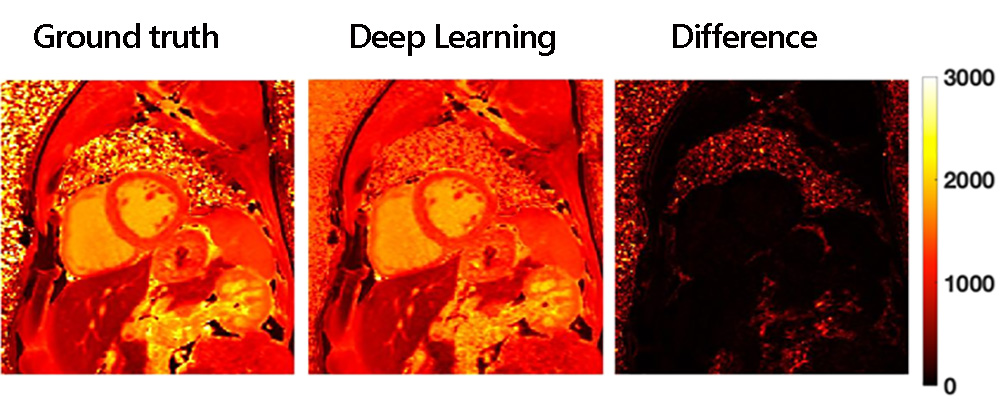
Results of the deep learning approach for rapid T1 mapping. The truth image required 11 heartbeats to create while the deep learning map only needed 5 heartbeats.
Right now, Adluru’s new application is a prototype and continues to be developed and tested. His hope is that it will be available to patients within a year.
From Faster Reconstruction to Faster Acquisition
Some MRI scans are quick but the reconstruction time is long. Other scans take a long time because a high level of detail needs to be acquired, although the reconstruction is fairly quick. A third paper from DiBella’s group shows how deep learning can dramatically speed up scan time for this second case.
Some years back, DiBella applied his compressed sensing acquisition technique developed for the heart muscle to MRI scans of the brain. He began working with a team of physicians and researchers including Jennifer Majersik, MD, the Chief of the Division of Vascular Neurology and Medical Director of the Stroke Center and Telestroke Services, as well as Lorie Richards, PhD, Chair and Associate Professor in the Division of Occupational Therapy in the College of Health. Majersik and Richards work with patients recovering from stroke, attempting to help them rehab and regain as much function as possible. MRI scans mapping the diffusion process of water molecules in the brain began to show promise for determining recovery after stroke. In fact, a type of MRI diffusion scan seemed to predict future motor function in patients. The only problem: the scan took 45 minutes. Thanks to compressed sensing and simultaneous multislice acquisition techniques, the group published a paper showing that a 13-minute MRI scan done within a week of having a stroke can be predictive of motor function 6 weeks later.

The stroke imaging with deep learning team (left to right): Ganesh Adluru, PhD, Ed DiBella, PhD, Jennifer Majersik, MD, Matthew Alexander, MD, Kazem Hashemizade, and Lorie Richards, PhD.
This three-fold gain in shortening the scan was great, but still not ideal for stroke patients, who undergo a battery of scans not just during the acute phase of the stroke but for months afterward. The team would like to add this “research” scan to any MRI scans being done in the course of follow-up, but 13 minutes on top of other scans that are being done is no small ask. Especially for stroke patients just days after their stroke who have a hard time getting positioned correctly in the scanner and staying still throughout. DiBella decided to apply AI to see if more gains could be made in shortening the scan time.
Diffusion imaging specifically looks at the white matter in the brain, which consists of dense bundles of axons that connect the brain’s functional areas. “The brain is essentially a thin sheath of grey matter, which are neurons, and the rest is the white matter that makes the connections between the neurons,” says Jeff Anderson, MD, PhD, neuroradiologist and researcher at UCAIR. In ischemic stroke, a blockage stops blood and oxygen flow to a region of the brain, resulting in dead white and grey matter. With hemorrhagic stroke, blood floods into the brain and damages white matter, pushing the connections out of their alignment. In both cases, the Diffusion MRI scan can measure the ability of water to move in the white matter and the direction in which it is moving. Clinicians can use the complex map of diffusion factors generated by the scan to determine which connections seem most damaged and which seem less damaged.
DiBella’s postdoctoral researcher, Eric Gibbons, PhD, set up a CNN and trained it on 41 data sets (1,459 total MRI scan “slices” from 15 stroke patients and 14 healthy volunteers). The network now had a good baseline of scenarios for how diffusion would manifest in stroke and healthy people. Next, he tested the network with data from 3 patients (365 MRI slices) – he compared their complete, baseline scans against the output from the CNN after he gave it undersampled data. The baseline scans measure how water diffuses in many different directions, and use different sensitivities to diffusion. 203 different diffusion-weighted images were collected as the gold standard. He found that they could get similar results by giving the network data from about one-tenth the number of diffusion-weighted images. By using data from just 24 directions of diffusion, the team’s deep learning network was able to provide valuable “orientation dispersion” data with just a 1.5-minute scan (see the paper here).
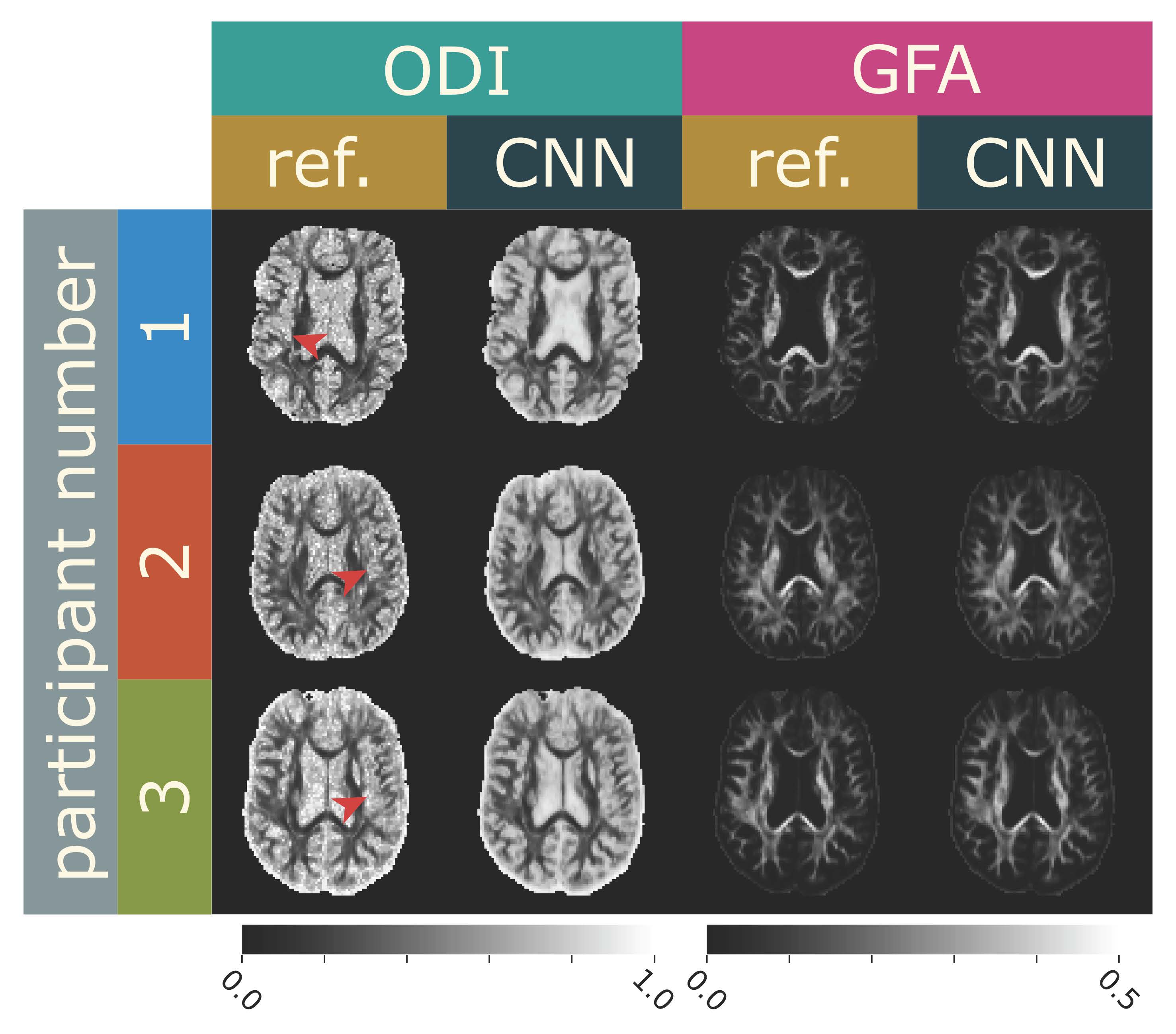
Comparison of brain scans from the 13-minute MRI scan that captures 203 diffusion directions (“ref”) and the scans from the 1.5-minute MRI scan using deep learning that only requires 24 diffusion directions.
“Our hope is that this type of scan using AI can become part of the standard workup for a patient recovering from a stroke,” says DiBella.
When AI Makes Sense
For scenarios like these, when the data demands of either image acquisition or image reconstruction are so great, it seems that using artificial intelligence makes a lot of sense. Computers can simply do complex calculations faster in trained convolutional networks than they can linearly (that is, “longhand,” one calculation after the next).
DiBella has several grant proposals in the works to continue his deep learning research, and several clinicians excited to partner with him. One of them, Matthew Alexander, MD, is an interventional neuroradiologist who treats acute stroke. “MRI is actually a more sensitive test for stroke in the early hours, detecting stroke when a CT can’t,” states Alexander. “I think it’s a great tool for clinicians because it can give us a number of quantitative factors beyond how tissues appear. And with AI speeding up scans, I hope that MRI becomes more user-friendly for clinicians and patients alike.”
DiBella is cautiously optimistic about the future of deep learning in imaging. “I’m definitely encouraged by what I’ve seen from some of these networks,” he says. “We hope to better understand the effectiveness of these approaches in the near future.”